Geo-grid processor
Converts geo-grid definitions of grid tiles or cells to regular bounding boxes or polygons which describe their shape. This is useful if there is a need to interact with the tile shapes as spatially indexable fields. For example the geotile field value "4/8/3" could be indexed as a string field, but that would not enable any spatial operations on it. Instead, convert it to the value "POLYGON ((0.0 40.979898069620134, 22.5 40.979898069620134, 22.5 55.77657301866769, 0.0 55.77657301866769, 0.0 40.979898069620134))", which can be indexed as a geo_shape field.
| Name | Required | Default | Description |
|---|---|---|---|
field |
yes | - | The field to interpret as a geo-tile. The field format is determined by the tile_type. |
tile_type |
yes | - | Three tile formats are understood: geohash, geotile and geohex. |
target_field |
no | field |
The field to assign the polygon shape to, by default field is updated in-place. |
parent_field |
no | - | If specified and a parent tile exists, save that tile address to this field. |
children_field |
no | - | If specified and children tiles exist, save those tile addresses to this field as an array of strings. |
non_children_field |
no | - | If specified and intersecting non-child tiles exist, save their addresses to this field as an array of strings. |
precision_field |
no | - | If specified, save the tile precision (zoom) as an integer to this field. |
ignore_missing |
no | - | If true and field does not exist, the processor quietly exits without modifying the document. |
target_format |
no | "GeoJSON" | Which format to save the generated polygon in. Either WKT or GeoJSON. |
description |
no | - | Description of the processor. Useful for describing the purpose of the processor or its configuration. |
if |
no | - | Conditionally execute the processor. See Conditionally run a processor. |
ignore_failure |
no | false |
Ignore failures for the processor. See Handling pipeline failures. |
on_failure |
no | - | Handle failures for the processor. See Handling pipeline failures. |
tag |
no | - | Identifier for the processor. Useful for debugging and metrics. |
To demonstrate the usage of this ingest processor, consider an index called geocells with a mapping for a field geocell of type geo_shape. In order to populate that index using geotile and geohex fields, define two ingest processors:
PUT geocells {
"mappings": {
"properties": {
"geocell": {
"type": "geo_shape"
}
}
}
}
PUT _ingest/pipeline/geotile2shape
{
"description": "translate rectangular z/x/y geotile to bounding box",
"processors": [
{
"geo_grid": {
"field": "geocell",
"tile_type": "geotile"
}
}
]
}
PUT _ingest/pipeline/geohex2shape
{
"description": "translate H3 cell to polygon",
"processors": [
{
"geo_grid": {
"field": "geocell",
"tile_type": "geohex",
"target_format": "wkt"
}
}
]
}
These two pipelines can be used to index documents into the geocells index. The geocell field will be the string version of either a rectangular tile with format z/x/y or an H3 cell address, depending on which ingest processor we use when indexing the document. The resulting geometry will be represented and indexed as a geo_shape field in either GeoJSON or the Well-Known Text format.
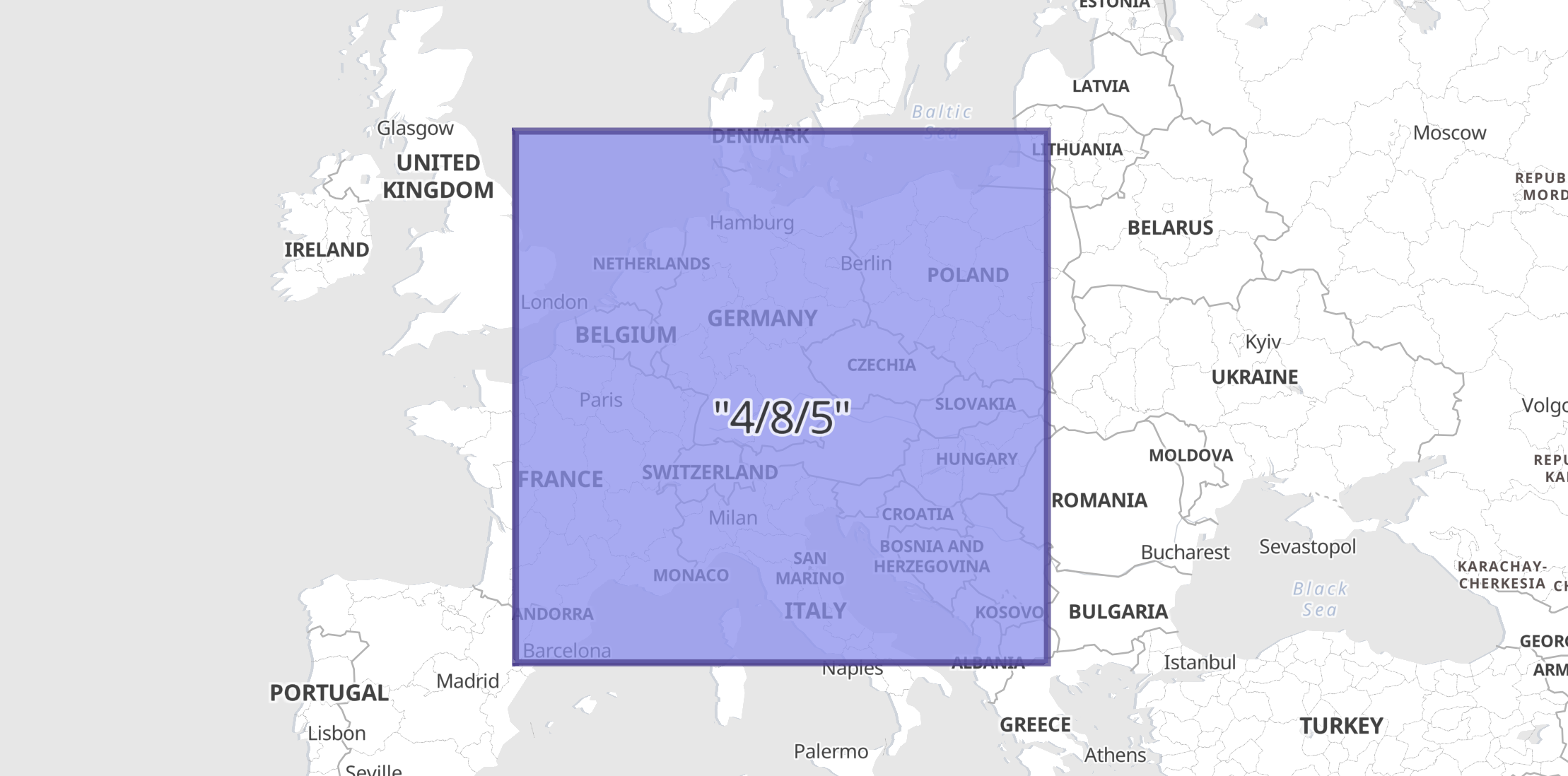
In this example a geocell field with a value defined in z/x/y format is indexed as a GeoJSON Envelope since the ingest-processor above was defined with default target_format.
PUT geocells/_doc/1?pipeline=geotile2shape {
"geocell": "4/8/5"
}
GET geocells/_doc/1
The response shows how the ingest-processor has replaced the geocell field with an indexable geo_shape:
{
"_index": "geocells",
"_id": "1",
"_version": 1,
"_seq_no": 0,
"_primary_term": 1,
"found": true,
"_source": {
"geocell": {
"type": "Envelope",
"coordinates": [
[ 0.0, 55.77657301866769 ],
[ 22.5, 40.979898069620134 ]
]
}
}
}
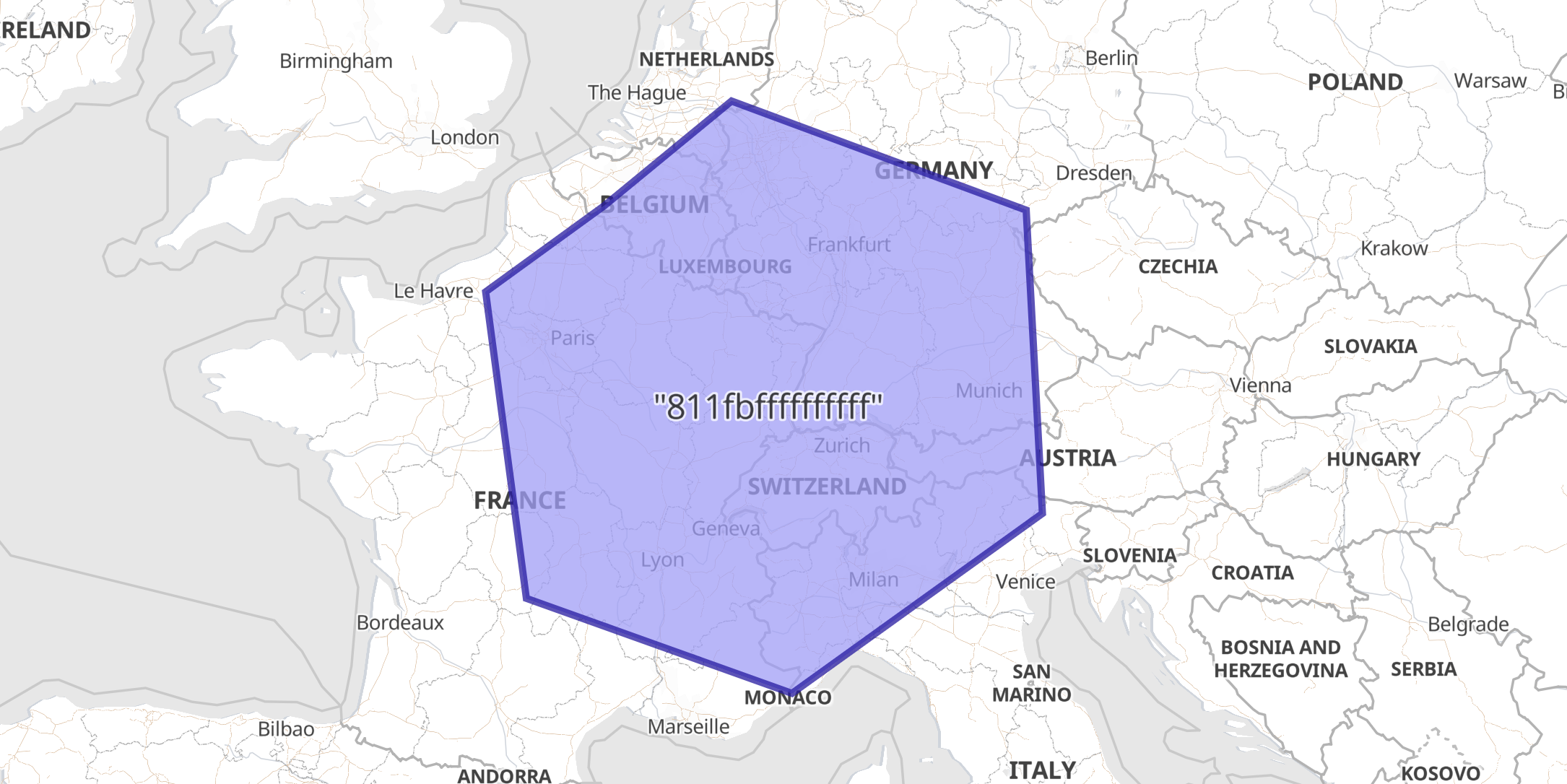
In this example a geocell field with an H3 string address is indexed as a WKT Polygon, since this ingest processor explicitly defined the target_format.
PUT geocells/_doc/1?pipeline=geohex2shape {
"geocell": "811fbffffffffff"
}
GET geocells/_doc/1
The response shows how the ingest-processor has replaced the geocell field with an indexable geo_shape:
{
"_index": "geocells",
"_id": "1",
"_version": 1,
"_seq_no": 0,
"_primary_term": 1,
"found": true,
"_source": {
"geocell": "POLYGON ((1.1885095294564962 49.470279179513454, 2.0265689212828875 45.18424864858389, 7.509948452934623 43.786609335802495, 12.6773177459836 46.40695743262768, 12.345747342333198 50.55427505169064, 6.259687012061477 51.964770150370896, 3.6300085578113794 50.610463307239115, 1.1885095294564962 49.470279179513454))"
}
}
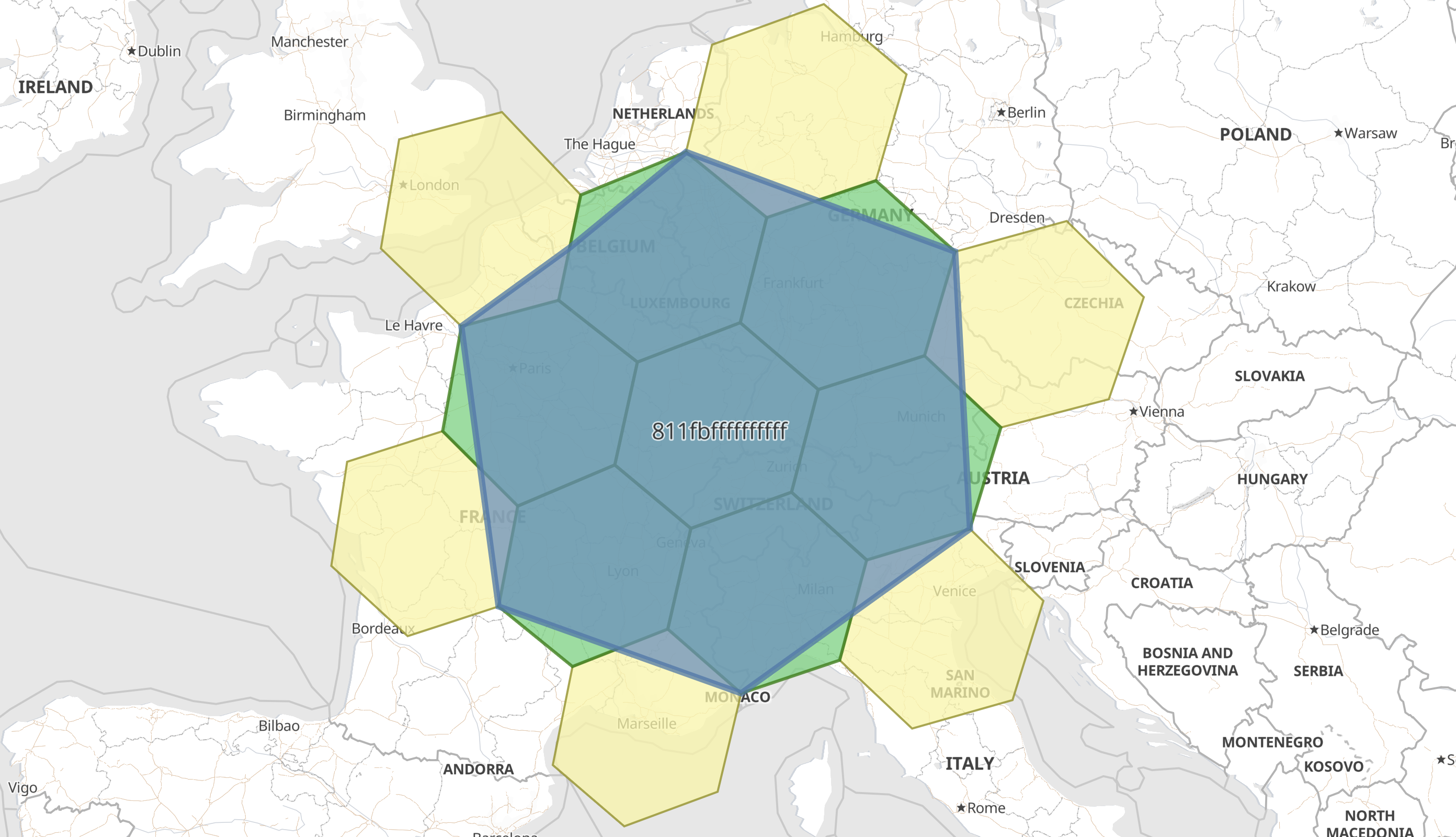
As described in geo_grid processor options, there are many other fields that can be set, which will enrich the information available. For example, with H3 tiles there are 7 child tiles, but only the first is fully contained by the parent. The remaining six are only partially overlapping the parent, and there exist a further six non-child tiles that overlap the parent. This can be investigated by adding parent and child additional fields to the ingest-processor:
PUT _ingest/pipeline/geohex2shape {
"description": "translate H3 cell to polygon with enriched fields",
"processors": [
{
"geo_grid": {
"description": "Ingest H3 cells like '811fbffffffffff' and create polygons",
"field": "geocell",
"tile_type": "geohex",
"target_format": "wkt",
"target_field": "shape",
"parent_field": "parent",
"children_field": "children",
"non_children_field": "nonChildren",
"precision_field": "precision"
}
}
]
}
Index the document to see a different result:
PUT geocells/_doc/1?pipeline=geohex2shape {
"geocell": "811fbffffffffff"
}
GET geocells/_doc/1
The response from this index request:
{
"_index": "geocells",
"_id": "1",
"_version": 1,
"_seq_no": 0,
"_primary_term": 1,
"found": true,
"_source": {
"parent": "801ffffffffffff",
"geocell": "811fbffffffffff",
"precision": 1,
"shape": "POLYGON ((1.1885095294564962 49.470279179513454, 2.0265689212828875 45.18424864858389, 7.509948452934623 43.786609335802495, 12.6773177459836 46.40695743262768, 12.345747342333198 50.55427505169064, 6.259687012061477 51.964770150370896, 3.6300085578113794 50.610463307239115, 1.1885095294564962 49.470279179513454))",
"children": [
"821f87fffffffff",
"821f8ffffffffff",
"821f97fffffffff",
"821f9ffffffffff",
"821fa7fffffffff",
"821faffffffffff",
"821fb7fffffffff"
],
"nonChildren": [
"821ea7fffffffff",
"82186ffffffffff",
"82396ffffffffff",
"821f17fffffffff",
"821e37fffffffff",
"82194ffffffffff"
]
}
}
This additional information will then enable, for example, creating a visualization of the H3 cell, its children and its intersecting non-children cells.